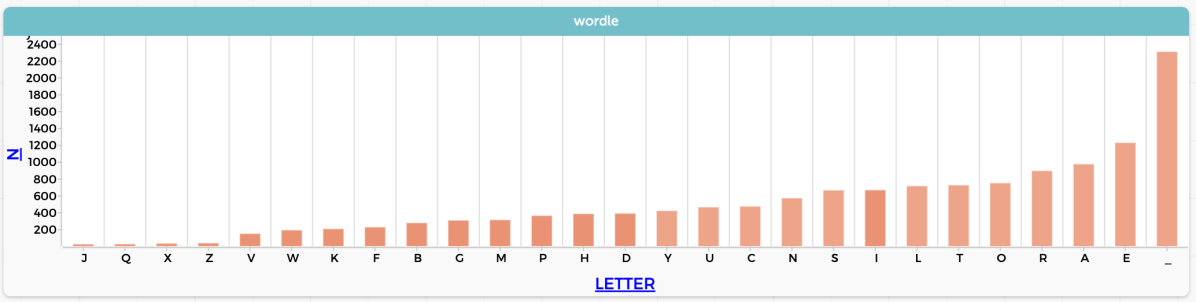
Long ago I promised a post (to come quickly; apparently I lied) about this topic.
If we have time-series data, we typically put time on the horizontal axis. But that’s not the only way to represent something that changes in time. There is an alternative that’s closely related to parametric equations and graphs.
Consider this typical physics-class situation: you have a weight on a spring moving up and down. You can plot its position as a function of time. For regular old Hooke-ian springs where , that will be a sinusoid.
Now plot the velocity as a function of time. That will also be a sinusoid, but shifted by , like this:
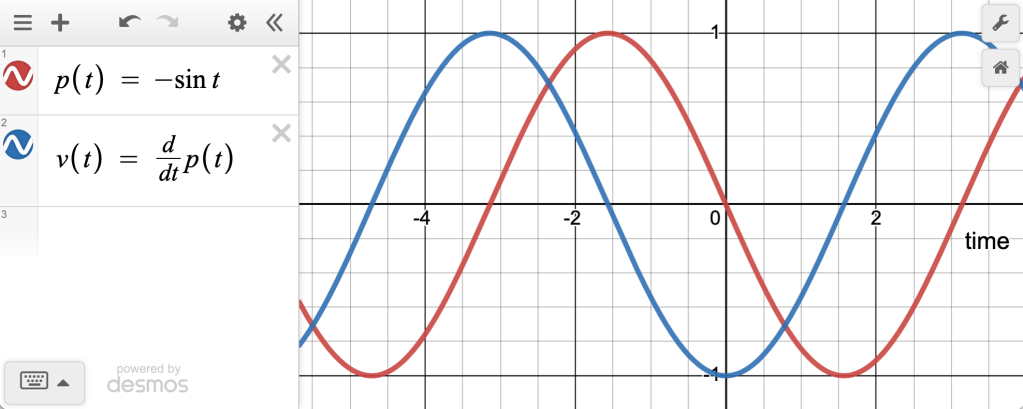
These are normal time-series functions, drawn with time on the horizontal axis. But as you may remember, you can instead plot them with the position on the axis and velocity on the
, making a new point for every value of
:
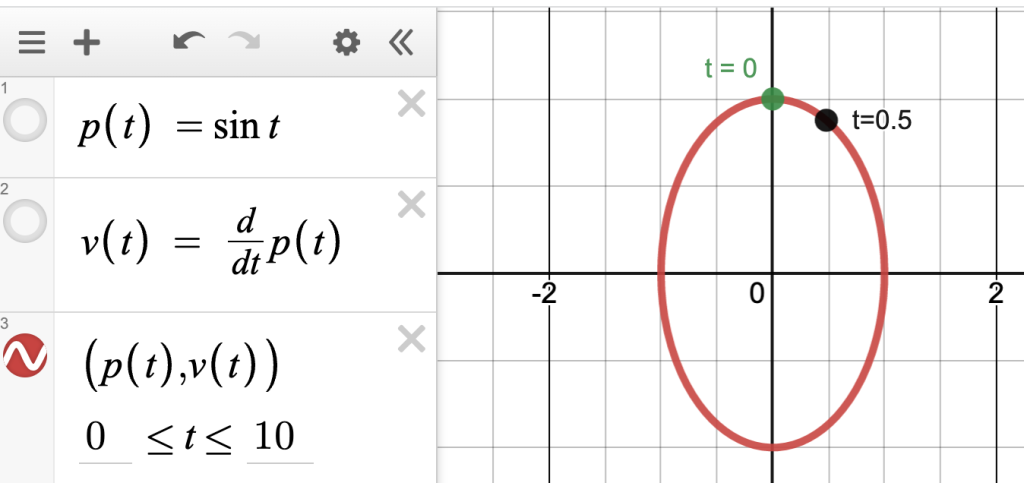
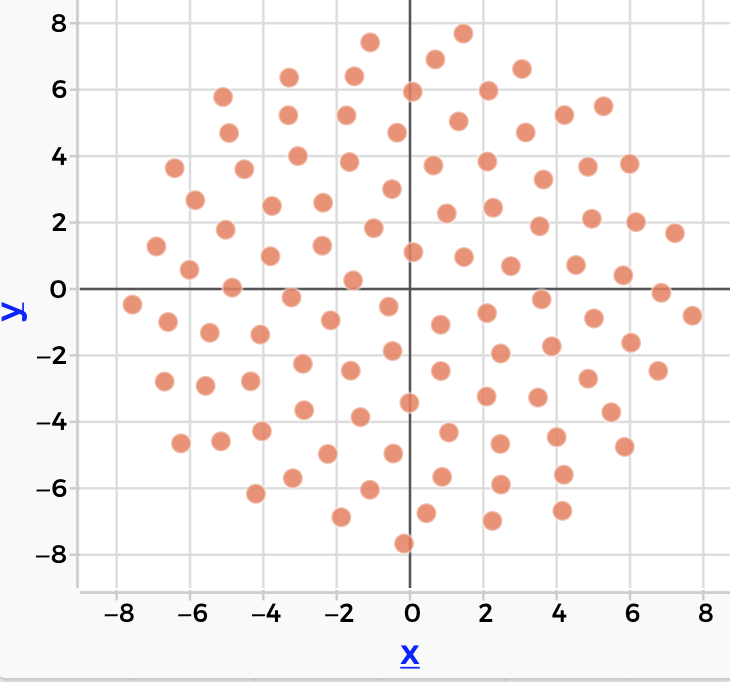
There are many cool things about this, like what happens when the frequency of is different from
, or when the delay is not 90°, and you get a wide variety of Lissajous figures. Or when you plot the velocity of a chaotic system against position…but that would be more of a digression than we want here.
Can we ever see this in real-world data, like, not from our imagination of a physics lab activity? Sure!
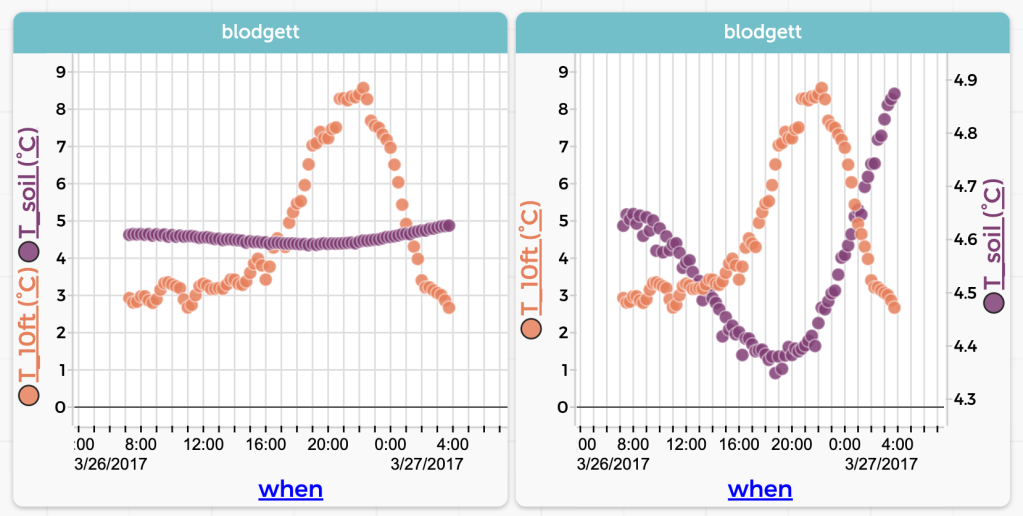
Here is one of my favorites, drawn from the “Blodgett” dataset available at codap.xyz. There we have air temperature in the Blodgett forest over the course of a year. We also have soil temperature. Here is one day of both plotted as traditional time series:
Now here is the same data plotted with soil temperature on the vertical axis and air on the horizontal. In this plot, time is going counterclockwise:
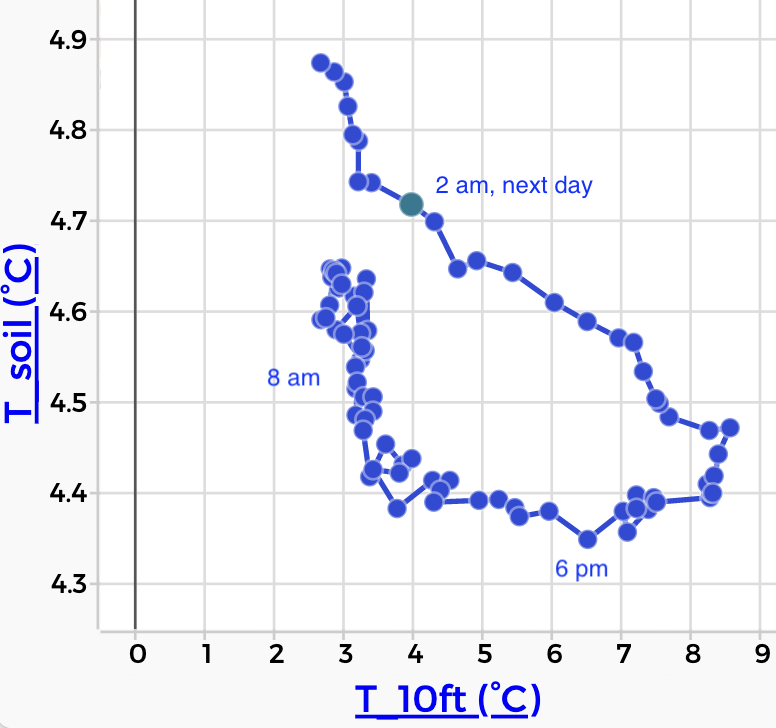
If you’re like me, that graph does not immediately shout what’s going on. But with some practice, we realize that the shape of the loop tells us about the delay between the air temperature and the soil temperature.
The point being, there are more ways in heaven and earth [Horatio] to display time series. Usually, of course, stick time on the horizontal. But be open to intriguing alternatives.